Everything You Need to Know about Vector Index Basics
This tutorial analyzes the components of a modern indexer before going over two simplest indexing strategies - flat indexing and inverted file (IVF) indexes.
Read the entire series
- Introduction to Unstructured Data
- What is a Vector Database and how does it work: Implementation, Optimization & Scaling for Production Applications
- Understanding Vector Databases: Compare Vector Databases, Vector Search Libraries, and Vector Search Plugins
- Introduction to Milvus Vector Database
- Milvus Quickstart: Install Milvus Vector Database in 5 Minutes
- Introduction to Vector Similarity Search
- Everything You Need to Know about Vector Index Basics
- Scalar Quantization and Product Quantization
- Hierarchical Navigable Small Worlds (HNSW)
- Approximate Nearest Neighbors Oh Yeah (Annoy)
- Choosing the Right Vector Index for Your Project
- DiskANN and the Vamana Algorithm
- Safeguard Data Integrity: Backup and Recovery in Vector Databases
- Dense Vectors in AI: Maximizing Data Potential in Machine Learning
- Integrating Vector Databases with Cloud Computing: A Strategic Solution to Modern Data Challenges
- A Beginner's Guide to Implementing Vector Databases
- Maintaining Data Integrity in Vector Databases
- From Rows and Columns to Vectors: The Evolutionary Journey of Database Technologies
- Decoding Softmax Activation Function
- Harnessing Product Quantization for Memory Efficiency in Vector Databases
- How to Spot Search Performance Bottleneck in Vector Databases
- Ensuring High Availability of Vector Databases
- Mastering Locality Sensitive Hashing: A Comprehensive Tutorial and Use Cases
- Vector Library vs Vector Database: Which One is Right for You?
- Maximizing GPT 4.x's Potential Through Fine-Tuning Techniques
- Deploying Vector Databases in Multi-Cloud Environments
- An Introduction to Vector Embeddings: What They Are and How to Use Them
In the previous tutorial, we went over a quick word embedding example to better understand the power of embeddings along with how they are stored and indexed in a vector database. This led to a brief overview of nearest neighbor search algorithms, a computing problem that involves finding the closest vector(s) to a query vector based on a selected distance metric.
Vector indexing involves mapping queries to a smaller subset of the vector space to enhance search efficiency. We then briefly discussed a couple of well-known methods for vector search: Vector search is a critical component of vector databases, but only as it relates to computation; vector databases are complex beasts that involve numerous moving components and layers of abstraction.
In this tutorial, we’ll analyze the components of a modern indexer before going over two of the simplest and most basic indexing strategies - flat indexing and inverted file indexes (IVF). Knowledge of these two indexing types will be critical as we progress into the next couple of tutorials.
Introduction
Vector indexing is a crucial component of vector databases, enabling efficient similarity searches in high-dimension datasets. With the increasing demand for fast and accurate search capabilities, understanding vector indexing and its various methods is essential for developers and data scientists. In this article, we will delve into the world of vector indexing, exploring its definition, importance, and common indexing methods.
What is Vector Indexing?
Definition of Vector Indexing
Vector indexing is a technique used to organize and store vector data in a way that enables fast and efficient similarity searches. It involves creating a data structure that maps vectors to their nearest neighbors, allowing for quick retrieval of similar vectors. Vector indexing is a critical component of vector databases, which are designed to handle high-dimensional data and provide fast query performance.
Vector indexing and Milvus
Milvus uses Facebook AI Similarity Search (FAISS) as one of the key indexing-building libraries, along with HnswlibAnnoy. As mentioned in the previous tutorial, Milvus builds on top of these libraries to provide a full-fledged database, complete with all the usual database features and a consistent user-level API. It is a good place to practice using a c++ vector index, as opposed to an R vector index, because FAISS is a library written in c++ with a Python interface.
If you’re already familiar with FAISS, many of the concepts introduced here and in the next couple of tutorials may already be familiar to you.
You may have noticed this after going through the previous tutorial, but there are, broadly speaking, four different types of vector search algorithms:
- hash-based indexing (e.g. locality-sensitive hashing),
- tree-based indexing (e.g. ANNOY),
- cluster-based or cluster indexing (e.g. product quantization), and
- graph-based indexing (e.g. Hierarchical navigable small world or HNSW, CAGRA).
Different types of algorithms work better for large datasets and varying types of vector data, but all of them help speed up the vector search process at the cost of a bit of accuracy/recall.
One key detail that often goes overlooked with vector search is the capability to combine many vector search algorithms together. Within a vector database, a full vector index is generally composed of three distinct components:
- an optional pre-processing step where vectors may be reduced or optimized prior to indexing,
- a required primary step which is the core algorithm used for indexing, and
- an optional secondary step where vectors may be quantized or hashed to further improve search speeds.
The first step simply prepares the vectors for indexing and search without actually building any data structure. The algorithm used here often depends on the application and the upstream vector generation method, but some commonly used ones include L2 normalization, dimensionality reduction, and zero padding. Most vector databases skip this step and leave pre-processing entirely up to the user in the application layer.
The primary algorithm is the only mandatory component and forms the crux of the vector index. The output of this step should be a data structure which holds all information necessary to conduct an efficient vector search. Tree-based and graph-based data structures are commonly used here, but a quantization algorithm or a hash based vector index such as product quantization or locality-sensitive hashing works as well. During this step, index creation is crucial, and selecting appropriate distance calculation methods ensures effective performance during query operations. Additionally, key parameters such as 'lists', 'probes', 'efConstruction', and 'efSearch' play significant roles in enhancing performance while managing memory usage. The representation and organization of data points are also essential to efficiently perform operations like k-nearest neighbor searches.
The secondary step reduces the total size of the index by mapping all floating point values in the dataset into lower-precision integer values, i.e. float64 -> int8 or float32 -> int8. This modification can both reduce the index size as well as improve search speeds, but generally at the cost of some precision. There are a couple different ways this can be done; we'll dive deeper into quantization and hashing in future tutorials.
Before diving too deep into more complex vector search algorithms, it pays take a brief look at linear search, also known as "flat" indexing.
A flat index is by and large the most basic indexing strategy, but arguably also the most overlooked. With flat indexing, we compare a query vector with every other vector in our database. In code, this would look something like this:
>>> query = np.random.normal(size=(128,))
>>> dataset = np.random.normal(size=(1000, 128))
>>> nearest = np.argmin(np.linalg.norm(dataset - query, axis=1))
>>> nearest
333
Note how the index is simply a flat data structure which is exactly the size of the dataset - no more and no less.
The first two lines of code creates a random query vector in addition to a 1000-element dataset of vectors. The third line then computes the distance (via np.linalg.norm) between all elements in the dataset and the nearest neighbors of the query vector before extracting the index of the minimum distance (via np.argmin). This gives us the array index of the nearest neighbor to the query vector, which we can then extract using dataset[nearest,:].
This is obviously the most naïve way to perform vector search, but it can work surprisingly well for small datasets, especially if you have an accelerator such as a GPU or FPGA to parallelize the search process on. The loop above, for example, runs in less than 0.5 milliseconds on an Intel i7-9750H CPU, which means that we can achieve a QPS of over 2000 with flat indexing on a six-core laptop CPU!
Our QPS drops to around 160 with 10k vectors and 16 with 100k vectors, with the >10x factor drop from 1000 vectors to 10k vectors likely being due to CPU cache size limitations. These numbers are still pretty good though. With all the talk nowadays about runtime complexity and horizontal scaling, remember the KISS principle for small applications and/or prototyping: Keep It Simple, Stupid.
Flat indexing is great, but it obviously doesn't scale. This is where data structures for vector search come into play. By trading off a bit of accuracy/recall for improved runtime, we can significantly improve both query speed and throughput. There are a lot of indexing strategies out there today, but one of the most commonly used ones is inverted file index (IVF).
Fancy name aside, IVF is actually fairly simple. An inverted file index reduces the overall search scope by arranging the entire dataset into partitions. All partitions are associated with a centroid, and every vector in the dataset is assigned to a partition that corresponds to its nearest centroid. The algorithm attempts to locate the nearest vectors within the same region as a query vector.
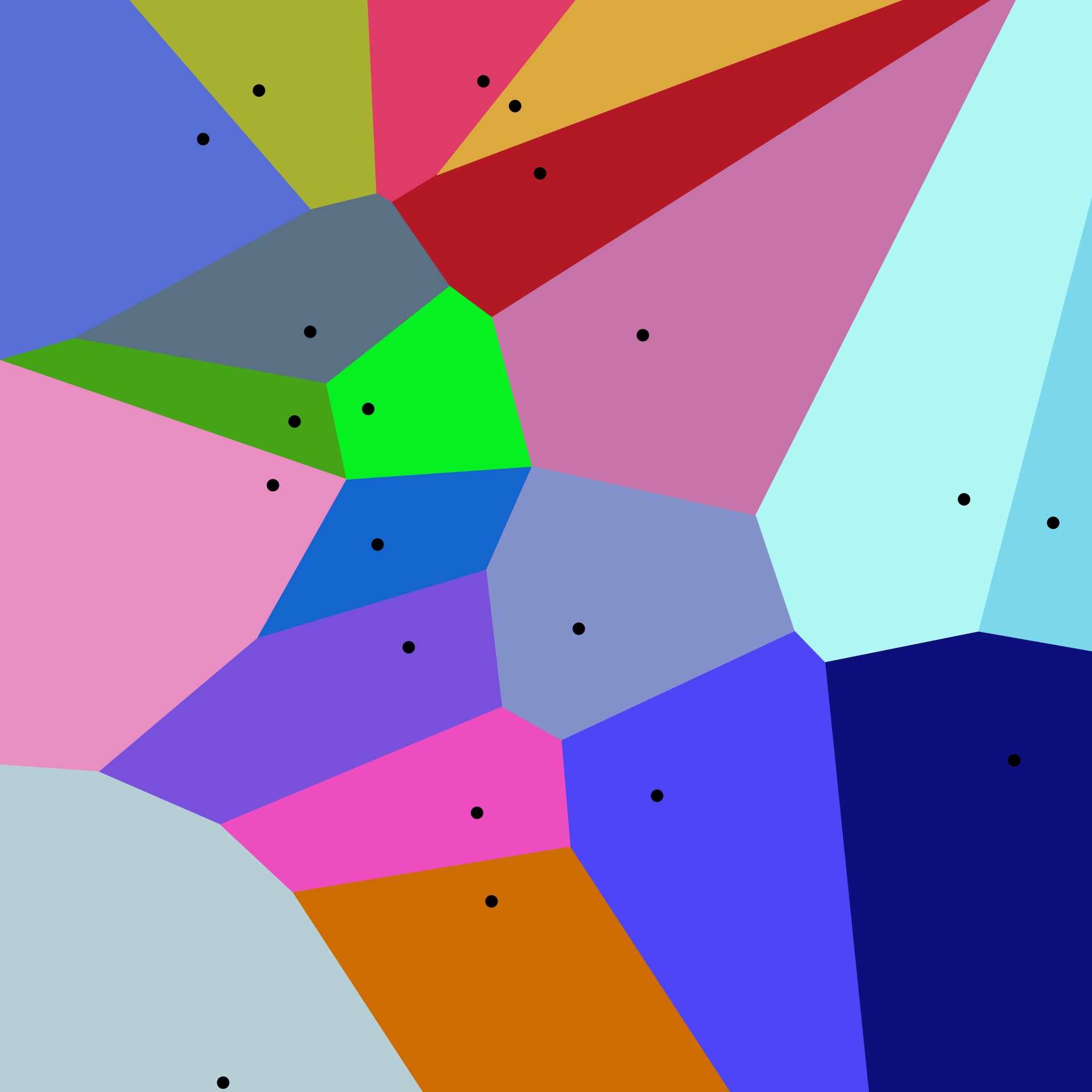 A two-dimensional Voronoi diagram. Image by Balu Ertl, CC BY-SA 4.0.
A two-dimensional Voronoi diagram. Image by Balu Ertl, CC BY-SA 4.0.
If you're familiar with FAISS, the above diagram might be familiar to you; it's called a Voronoi diagram and visually illustrates this cluster index assignment, albeit in only two dimensions. There are a total of 20 cells (clusters), with the centroid for each cluster displayed as a black dot. All points in a dataset will fall into one of these 20 regions.
Cluster centroids are usually determined with a clustering algorithm called k-means. K-means is an interative algorithm that works by first randomly selecting a set of K points as clusters. At every iteration, all points in the dataset of vectors are assigned to its nearest centroid, and all centroids are then updated to the mean of each cluster. This process then continues until convergence - a process known as expectation-maximazation for folks familiar with statistics.
Armed with this knowledge, let's use k-means to "automagically" determine centroids for IVF. For this, we'll use scipy's kmeans2 implementation:
>>> import numpy as np
>>> from scipy.cluster.vq import kmeans2
>>> num_part = 16 # number of IVF partitions
>>> dataset = np.random.normal(size=(1000, 128))
>>> (centroids, assignments) = kmeans2(dataset, num_part, iter=32)
>>> centroids.shape
(16, 128)
>>> indexes.shape
(1000,)
centroids now contains all num_part (in FAISS, this parameter is called nlist) centroids for our dataset, while assignments contains ID of the centroid/cluster that is closest to each vector. We can verify this as follows:
>>> test = [np.argmin(np.linalg.norm(vec - centroids, axis=1)) for vec in dataset]
>>> np.all(test == assignments)
True
We'll now need to create the inverted file index by correlating each centroid with a list of vectors within the cluster:
>>> index = [[] for _ in range(num_part)]
>>> for n, k in enumerate(assignments):
... index[k].append(n) # the nth vector gets added to the kth cluster
...
The code above first creates a list of lists, with the outermost layer corresponding to the number of inverted file index partitions. The for loop then loops through all assignments (i.e. which partition each vector belongs to) and populates the index.
With the index in place, we can now restrict the overall search space by searching only the nearest cluster:
>>> query = np.random.normal(size=(128,))
>>> c = np.argmin(np.linalg.norm(centroids - query, axis=1)) # find the nearest partition
>>> nearest = np.argmin(np.linalg.norm(dataset[index[c]] - query, axis=1)) # find nearest neighbor
>>> nearest
333
With an num_part value of 16 and a dataset size of 100k, we get around 150 QPS using the same hardware as before (Intel i7-9750H CPU). Bumping num_part to 64 nets us a whopping 650 QPS.
Note that it’s often pragmatic to extend our search beyond just the nearest cluster, especially for high-dimensional data (for those familiar with FAISS, this corresponds to the `nprobe` parameter when creating an inverted file index). This is largely due to the curse of dimensionality, where each partition has a significantly larger number of edges when compared with similar data in two or three dimensions. There’s no good rule of thumb for a good value of `nprobe` to use - rather, it’s helpful to first experiment with your data to see the speed versus accuracy/recall tradeoffs.
And that’s it for the inverted file index! Not too bad, right?
In this tutorial, we looked at the three individual components of a vector index along with two of the most commonly used methods - flat indexing and the inverted file index. These are two of the most basic strategies, and we’ll use them as a launchpad for further deep dives into more complex vector indices.
In the next tutorial, we’ll continue our deep dive into indexing strategies with scalar quantization (SQ) and product quantization (PQ) - two popular quantization strategies available to Milvus users. See you in the next tutorial!
All code for this tutorial is freely available on Github: https://github.com/fzliu/vector-search.
Common Indexing Methods
Flat Indexing
Flat indexing is a simple and straightforward indexing method that stores vectors as is, without any modifications. It is the most basic indexing strategy, but also the most overlooked. Flat indexing provides perfect accuracy but is slow, making it suitable for small datasets where search speed is reasonable. However, as the dataset size increases, flat indexing becomes impractical due to its slow search speed.
In the next section, we will explore other common indexing methods, including Locality Sensitive Hashing (LSH), Inverted File (IVF), and Hierarchical Navigable Small Worlds (HNSW). These methods offer a trade-off between search speed and accuracy, making them suitable for different use cases and dataset sizes.
Start Free, Scale Easily
Try the fully-managed vector database built for your GenAI applications.
Try Zilliz Cloud for FreeKeep Reading

Introduction to Milvus Vector Database
Zilliz tells the story about building the world's very first open-source vector database, Milvus. The start, the pivotal, and the future.

DiskANN and the Vamana Algorithm
Dive into DiskANN, a graph-based vector index, and Vamana, the core data structure behind DiskANN.

Ensuring High Availability of Vector Databases
Ensuring high availability is crucial for the operation of vector databases, especially in applications where downtime translates directly into lost productivity and revenue.
