How to Select Index Parameters for IVF Index

In Best Practices for Milvus Configuration, some best practices for Milvus 0.6.0 configuration were introduced. In this article, we will also introduce some best practices for setting key parameters in Milvus clients for operations including creating a table, creating indexes, and searching. These parameters can affect search performance.
1. index_file_size
When creating a table, the index_file_size parameter is used to specify the size, in MB, of a single file for data storage. The default is 1024. When vector data is being imported, Milvus incrementally combines data into files. When the file size reaches index_file_size, this file does not accept new data and Milvus saves new data to another file. These are all raw data files. When an index is created, Milvus generates an index file for each raw data file. For the IVFLAT index type, the index file size approximately equals to the size of the corresponding raw data file. For the SQ8 index, the size of an index file is approximately 30 percent of the corresponding raw data file.
During a search, Milvus searches each index file one by one. Per our experience, when index_file_size changes from 1024 to 2048, the search performance improves by 30 percent to 50 percent. However, if the value is too large, large files may fail to be loaded to GPU memory (or even CPU memory). For example, if GPU memory is 2 GB and index_file_size is 3 GB, the index file cannot be loaded to GPU memory. Usually, we set index_file_size to 1024 MB or 2048 MB.
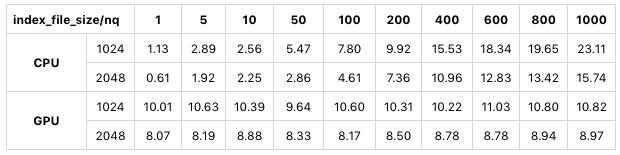
The following table shows a test using sift50m for index_file_size. The index type is SQ8.
 1-sift50m-test-results-milvus.
1-sift50m-test-results-milvus.
We can see that in CPU mode and GPU mode, when index_file_size is 2048 MB instead of 1024 MB, the search performance significantly improves.
2. nlist and nprobe
The nlist parameter is used for index creating and the nprobe parameter is used for searching. IVFLAT and SQ8 both use clustering algorithms to split a large number of vectors into clusters, or buckets. nlist is the number of buckets during clustering.
When searching using indexes, the first step is to find a certain number of buckets closest to the target vector and the second step is to find the most similar k vectors from the buckets by vector distance. nprobe is the number of buckets in step one.
Generally, increasing nlist leads to more buckets and fewer vectors in a bucket during clustering. As a result, the computation load decreases and search performance improves. However, with fewer vectors for similarity comparison, the correct result might be missed.
Increasing nprobe leads to more buckets to search. As a result, the computation load increases and search performance deteriorates, but search precision improves. The situation may differ per datasets with different distributions. You should also consider the size of the dataset when setting nlist and nprobe. Generally, it is recommended that nlist can be 4 * sqrt(n), where n is the total number of vectors. As for nprobe, you must make a trade-off between precision and efficiency and the best way is to determine the value through trial and error.
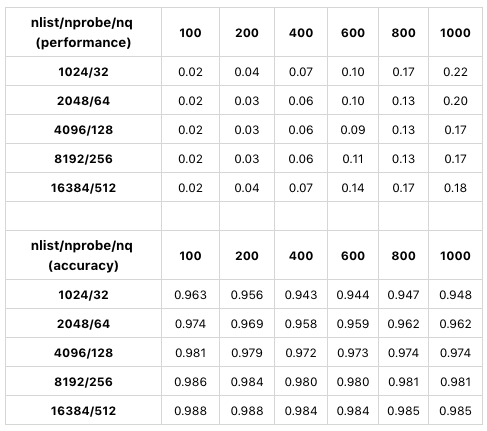
The following table shows a test using sift50m for nlist and nprobe. The index type is SQ8.
 sq8-index-test-sift50m.
sq8-index-test-sift50m.
The table compares search performance and precision using different values of nlist/nprobe. Only GPU results are displayed because CPU and GPU tests have similar results. In this test, as the values of nlist/nprobe increase by the same percentage, search precision also increase. When nlist = 4096 and nprobe is 128, Milvus has the best search performance. In conclusion, when determining the values for nlist and nprobe, you must make a trade-off between performance and precision with consideration to different datasets and requirements.
Summary
index_file_size: When the data size is greater than index_file_size, the greater the value of index_file_size, the better the search performance.
nlist and nprobe:You must make a trade-off between performance and precision.
Start Free, Scale Easily
Try the fully-managed vector database built for your GenAI applications.
Try Zilliz Cloud for FreeKeep Reading

How to Improve Retrieval Quality for Japanese Text with Sudachi, Milvus/Zilliz, and AWS Bedrock
Learn how Sudachi normalization and Milvus/Zilliz hybrid search improve Japanese RAG accuracy with BM25 + vector fusion, AWS Bedrock embeddings, and practical code examples.

Zilliz Cloud Audit Logs Goes GA: Security, Compliance, and Transparency at Scale
Zilliz Cloud Audit Logs are GA—delivering security, compliance, and transparency at scale with real-time visibility and enterprise-ready audit trails.
The Great AI Agent Protocol Race: Function Calling vs. MCP vs. A2A
Compare Function Calling, MCP, and A2A protocols for AI agents. Learn which standard best fits your development needs and future-proof your applications.
