DiskANN: A Disk-based ANNS Solution with High Recall and High QPS on Billion-scale Dataset
“DiskANN: Fast Accurate Billion-point Nearest Neighbor Search on a Single Node” is a paper published on NeurIPS in 2019. The paper introduces a state-of-the-art method to perform index building and search on the billion-scale dataset using a single machine with only 64GB of RAM and a large enough SSD. Moreover, it satisfies the three requirements of ANNS (Approximate Nearest Neighbor Search) on the large-scale dataset: high recall, low latency, and high density (number of nodes in a single machine). This method builds a graph-based index on a billion-scale dataset SIFT-1B using a single machine with 64GB of RAM and a 16-core CPU, reaching 5000 QPS (queries per second) at over 95 % recall@1, and the average latency lower than 3ms.
Authors
Suhas Jayaram Subramanya: Former employee of Microsoft India Research Institute, doctoral student of CMU. The main research interests are high-performance computing and machine learning algorithms for large-scale data.
Devvrit: Graduate Research Assistant at The University of Texas at Austin. His research interests are theoretical computer science, machine learning, and deep learning.
Rohan Kadekodi: A doctoral student at the University of Texas. His research direction is system and storage, mainly including persistent storage, file system, and kV storage.
Ravishankar Krishaswamy: Microsoft Indian research institute principal researcher. Doctor of CMU. The research direction is the approximation algorithm based on graph and clustering.
Harsha Vardhan Simhadri: Microsoft Indian research institute principal researcher. Doctor of CMU. In the past, he studied parallel algorithms and runtime systems. Now his main work is to develop new algorithms and write programming models.
Motivations
Most of the mainstream ANNS algorithms make some trade-offs among index building performance, search performance, and recall. Graph-based algorithms such as HNSW and NSG are state-of-art methods in terms of search performance and recall at present. Since the memory-resident graph-based indexing method occupies too much memory, it is relatively difficult to index and search a large-scale dataset using a single machine with limited memory resources.
Many applications require quick responses of Euclidean distance-based ANNS on the billion-scale dataset. Below are two major solutions:
Inverted index + quantization: to cluster the dataset into M partitions and compress the dataset using quantization schemes such PQ (Product Quantization). This solution produces low recall because of a loss of precision caused by data compression. Increasing the topk helps improve the recall while the QPS would drop correspondingly.
Divide and index: to divide the dataset into several disjoint shards and build an in-memory index for each shard. When query requests come, the search will be performed on indexes of each shard and the results will be returned after merging. This solution causes the over-expansion of the dataset scale, and thus more machines are needed because of the restriction of memory resources in a single machine, leading to low QPS.
Both solutions mentioned above are limited by the memory restriction of a single machine. This paper proposes the design of an SSD-resident indexing mechanism to solve this problem. The challenge of SSD-resident indexing is to reduce the number of random disk access and the number of requests for disk access.
Contributions
This paper presents an SSD-resident ANNS scheme called DiskANN, which can effectively support search on large-scale datasets. This scheme is based on a graph-based algorithm presented in this paper: Vamana. Contributions of this paper include:
DiskANN can index and search a billion-scale dataset of over 100 dimensions on a single machine with 64GB RAM, providing over 95% recall@1 with latencies under 5 milliseconds.
A new graph-based algorithm called Vamana with a smaller search radius than those of NSG and HNSW was proposed to minimize the number of disk access.
Vamana can work in memory and its performance is not slower than NSG and HNSW.
Smaller Vamana indexes built on overlapping partitions of the large dataset can be merged into one graph without losing connectivity.
Vamana can be combined with quantization schemes such as PQ. The graph structure and the original data are stored on the disk while compressed data is kept in memory.
Vamana
This algorithm is similar to the idea of NSG[2][4] (for those who don't understand NSG, please refer to Reference [2], and if you do not want to read papers, you can refer to Reference [4]). Their main difference lies in the trimming strategy. To be precise, a switch alpha has been added to the NSG's trimming strategy. The main idea of the NSG trimming strategy is that the choice of neighbors of the target point is as diverse as possible. If the new neighbor is closer to a neighbor of the target point than the target point, we do not need to add this point to the neighbor point set. In other words, for each neighbor of the target point, there can be no other neighbor points within the surrounding radius dist (target point, neighbor point). This trimming strategy effectively controls the out-degree of graph, and is relatively radical. It reduces the memory footprint of the index, improves the search speed, but also reduces the search accuracy. Vamana's trimming strategy is to freely control the scale of trimming through the parameter alpha. The working principle is to multiply the dist (a neighbor point, candidate point) in the trimming condition with a parameter alpha (not less than 1). Only when the dist (target point, a certain candidate point) is greater than the enlarged reference distance is the trimming strategy adopted, increasing the tolerance of mutual exclusion between neighbors of the target point.
Vamana's indexing process is relatively simple:
Initialize a random graph;
Calculate the starting point, which is similar to the navigation point of NSG. First, find the global centroid, and then find the point closest to the global centroid as the navigation point. The difference between Vamana and NSG is that the input of NSG is already a nearest neighbor graph, so users can simply do an approximate nearest neighbor search on the centroid point directly on the initial neighbor graph. However, Vamana initializes a random nearest neighbor graph, thus users cannot conduct approximate search directly on the random graph. They need to do a global comparison to get a navigation point as the starting point of subsequent iterations. The purpose of this point is to minimize the average search radius;
Perform Approximate Nearest Neighbor Search on each point based on the initialized random neighbor graph and the search starting point determined in step 2, make all points on the search path the candidate neighbor sets, and execute the edge trimming strategy using alpha = 1. Similar to that of NSG, selecting the point set on the search path starting from the navigation point as the candidate neighbor set will increase some long edges and effectively reduce the search radius.
Adjust alpha > 1 (the paper recommends 1.2) and repeat step 3. Whereas step 3 is based on a random nearest neighbor graph, the graph is low-quality after the first iteration. Therefore, another iteration is needed to improve the graph quality, which is very important to the recall rate.
This paper compares the three graph indexes, i.e. Vamana, NSG, and HNSW. In terms of indexing and query performance, Vamana and NSG are relatively close, and both outmatch HNSW slightly. Refer to the Experiment section below for the data.
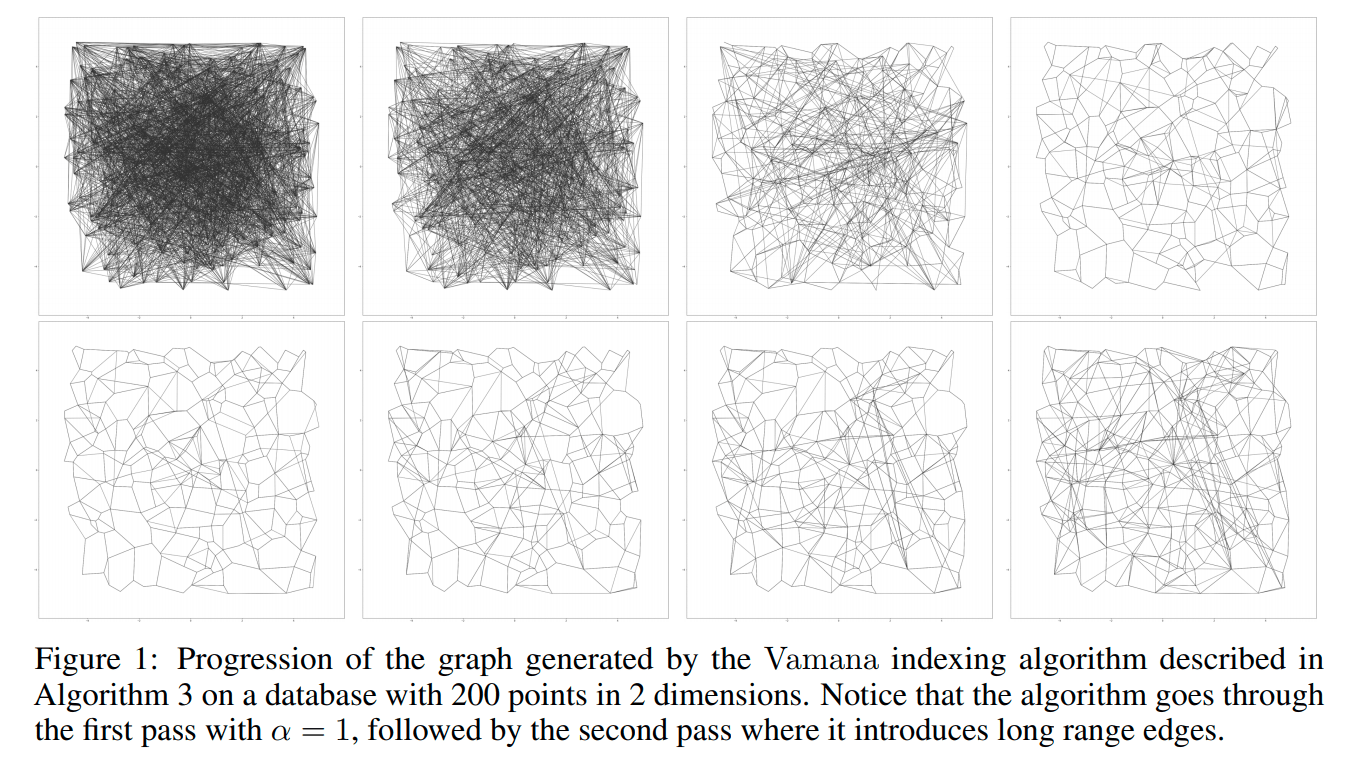 Figure 1.
Figure 1.
To visualize the building process of Vamana index, the paper provides a graph, in which 200 two-dimensional points are used to simulate two rounds of iteration. The first row uses alpha = 1 to trim the edges. It can be seen that the trimming strategy is relatively radical, and a large number of edges are trimmed. After increasing the value alpha and loosening the trimming conditions, a lot of edges are obviously added back. In the final graph, quite some long edges are added. It can effectively reduce the search radius.
DiskANN
A personal computer with only 64GB of memory would not even hold a billion pieces of raw data, let alone the index built on them. There are two challenges ahead: 1. How to index such a large-scale data set with limited memory resources? 2. How to calculate the distance when searching if the original data cannot be loaded in memory?
The paper proposed the following solutions:
For the first challenge: first, divide the data into k clusters using k-means, and then allocate each point into nearest i clusters. Generally, 2 is enough for the number i. Build a memory-based Vamana index for each cluster, and finally merge k Vamana indexes into one.
For the second challenge: build index on the original vectors and query compressed vectors. Building indexes on the original vector ensures the quality of the graph, while the compressed vector can be loaded in the memory for coarse-grained search. Although searching with the compressed vectors may cause a loss of accuracy, the general direction will be correct as long as the quality of the graph is high enough. The final distance result will be calculated using the original vector.
The index layout of DiskANN is similar to those of the general graph indexes. The neighbor set of each point and the original vector data are stored together. This makes better use of the locality of the data.
As mentioned earlier, if the index data is stored on the SSD, the number of disk accesses and the disk read and write requests must be reduced as much as possible to ensure low search delay. Therefore DiskANN proposes two optimization strategies:
Cache hotspot: cache all points within C jumps from the starting point in memory. The value of C is better set within 3 to 4.
Beam search: Simply put, it is to preload the neighbor information. When searching for point p, the neighbor point of p needs to be loaded from the disk if it is not in memory. Since a small amount of SSD random access operation takes about the same time as an SSD single-sector access operation, the neighbor information of W non-accessed points can be loaded at a time. W cannot be set too large or small. A large W will waste computing resources and SSD bandwidth, while a small one will increase the search delay.
Experiment
The experiment consists of three groups:
Comparison among memory-based indexes: Vamana VS. NSG VS. HNSW
Data sets: SIFT1M (128 dimensions), GIST1M (960 dimensions), DEEP1M (96 dimensions) and a 1M data set randomly sampled from DEEP1B.
Index parameters (all data sets use the same set of parameters):
HNSW:M = 128, efc = 512.
Vamana: R = 70, L = 75, alpha = 1.2.
NSG: R = 60, L = 70, C= 500.
The search parameters are not provided in the paper, which may be consistent with the indexing parameters. For the parameter selection, the parameters of NSG mentioned in the article are based on the parameters listed in the GitHub repository of NSG to select the group with better performance. Vamana and NSG are relatively close, so the parameters are also set close. However, the reason of HNSW parameters selection is not given. We believe that the parameter M of HNSW is set relatively large. It might lead to a less convincing comparison between graph-based indexes if their out-degrees are not set at the same level.
Under the above indexing parameters, the indexing time of Vamana, HNSW, and NSG are 129s, 219s, and 480s respectively. The NSG indexing time includes the time to construct the initial neighbor graph with EFANN [3].
Recall-QPS curve:
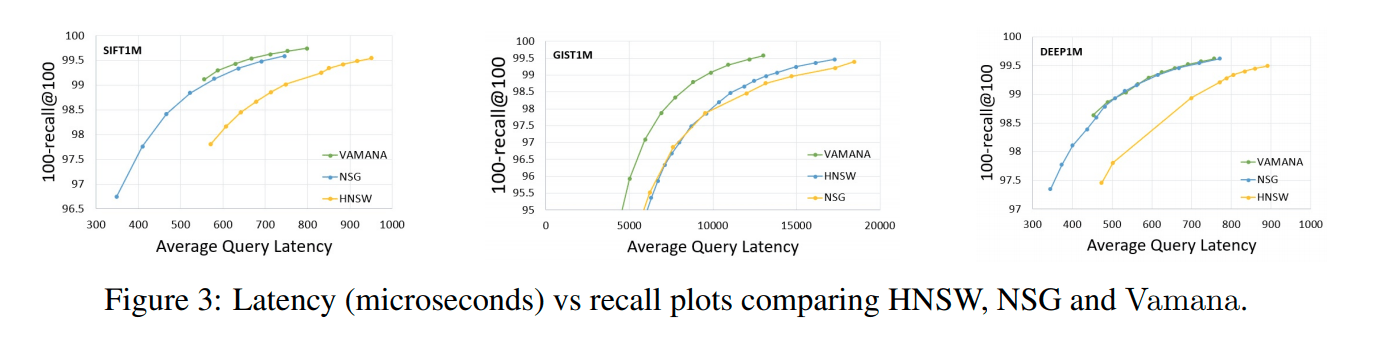 Figure 2.
Figure 2.
It can be seen from Figure 3 that Vamana has an excellent performance on the three data sets, similar to NSG and slightly better than HNSW.
Comparison of search radius:
From Figure 2.c, we can see that Vamana has the shortest average search path under the same recall rate compared to those of NSG and HNSW.
Comparison between a one-time built index and a large merged index
Data set: SIFT1B
The one-time built index parameters: L = 50, R = 128, alpha = 1.2. After running for 2 days on a 1800G DDR3 machine, the peak memory is about 1100 G, and the average out-degree is 113.9.
Indexing procedure based on the merging:
Train 40 clusters on the dataset using kmeans;
Each point is distributed into the nearest 2 clusters;
Build a Vamana index with L = 50, R = 64, and alpha = 1.2 for each cluster;
Merge the indexes of each cluster.
This index generated a 384GB index with an average out-of-degree of 92.1. This index ran for 5 days on a 64GB DDR4 machine.
The comparison results are as follows (Figure 2a):
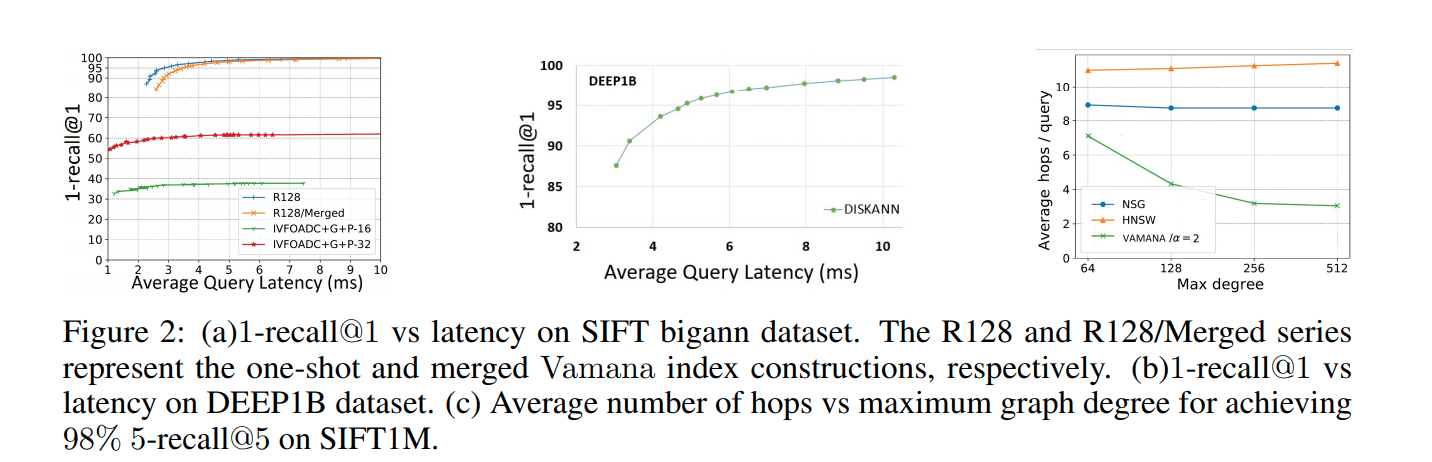 Figure 3.
Figure 3.
In conclusion:
The one-time built index is significantly better than the merging-based index;
The merging-based index is also excellent;
The merging-based indexing scheme is also applicable to the DEEP1B data set (Figure 2b).
Disk-based index: DiskANN VS. FAISS VS. IVF-OADC+G+P
IVFOADC+G+P is an algorithm proposed in Reference [5].
This paper only compares DiskANN with IVFOADC+G+P, since the reference [5] has proved that IVFOADC+G+P is better than FAISS. In addition, FAISS requires GPU resources, which are not supported by all platforms.
IVF-OADC+G+P seems to be a combination of HNSW and IVF-PQ. It determines clusters using HNSW, and performs search by adding some pruning strategies to the target cluster.
The result is in Figure 2a. The 16 and 32 in the figure are the codebook size. The dataset is SIFT1B, quantified by OPQ.
Code implementation details
The source code of DiskANN is open-sourced on https://github.com/microsoft/DiskANN
In January 2021, the source code of the disk solution was open-sourced.
The following mainly introduces the indexing process and the search process.
Index building
There are 8 parameters for building index:
data_type: options include float/int8/uint8.
data_file.bin: The original data binary file. The first two integers in the file respectively represent the total number n of the dataset vector and the vector dimension dim. The last n * dim * sizeof(data_type) bytes are continuous vector data.
index_prefix_path: The path prefix of the output file. After the index is built, several index-related files will be generated. This parameter is the common prefix of the directory where they are stored.
R: The maximum out-degree of the global index.
L: The parameter L of Vamana index, the upper bound of the candidate set size.
B: The memory threshold when querying. It controls the PQ codebook size, in GB.
M: The memory threshold when building an index. It determines the size of the fragment, in GB.
T: The number of threads.
Indexing process (entry function: aux_utils.cpp::build_disk_index):
Generate various output file names according to index_prefix_path.
Parameter check.
Read the meta of data_file.bin to get n and dim. Determine the codebook subspace number m of PQ according to B and n.
generate_pq_pivots: Sample the center point of the PQ training set using the sampling rate of p = 1500000/n uniformly to train PQ globally.
generate_pq_data_from_pivots: Generate global PQ codebook, and save the center point and codebook separately.
build_merged_vamana_index: slice the original data set, build Vamana indexes in segments, and finally merge the indexes in one.
partition_with_ram_budget: Determine the number of fragments k according to the parameter M. Sample the data set using kmeans, disributing each point to two nearest clusters. Fragment the dataset, and each fragment produces two files: a data file and an ID file. The ID file and the data file correspond to each other, and each ID in the ID file corresponds to a vector in the data file. The ID are obtained by numbering each vector of the original data from 0 to n-1. The ID is relatively important and is related to the merge.
Globally uniformly sample the training set with a sampling rate of 1500000 / n;
Initialize num_parts = 3. Iterate from 3:
- Do num_parts-means++ on the training set in step i;
- Use a sampling rate of 0.01 to sample a test set uniformly globally, and divide the test set into the nearest 2 clusters;
- Count the number of points in each cluster and divide it by the sampling rate to estimate the number of points in each cluster;
- Estimate the memory required by the largest cluster in step 3 according to the Vamana index size, if it does not exceed the parameter M, go on to step iii, otherwise num_parts ++ go back to step 2;
Divide the original data set into num_parts group files, each group of files includes fragmented data files and ID files corresponding to the fragmented data.
Create Vamana indexes separately for all the slices in step a and save them to disk;
merge_shards: merge num_parts shard Vamana into a global index:
Read the ID file of num_parts fragments into idmap. This idmap is equivalent to establishing a forward mapping of fragment->id;
Establish a reverse mapping of id-> fragments according to idmap, and know which two fragments each vector is in;
Use a reader with 1GB cache to open num_parts slice Vamana indexes, and use a writer with 1GB cache to open the output file, ready to merge;
Place num_parts navigation points of Vamana index into the center point file, which will be used when searching;
Start merging according to ID from small to large, read the neighbor point set of each original vector in each fragment in turn according to the reverse mapping, deduplicate, shuffle, truncate, and write to the output file. Because the slicing was originally globally ordered, and now the merging is also in order, so the ID in the final flushed index and the ID of the original data are one-to-one correspondence.
Delete temporary files, including fragment files, fragment indexes, and fragment ID files.
7.create_disk_layout: The global index generated in step 6 only has only a compact adjacency table. This step is to align the index. The adjacency table and the original data are stored together. When searching, load the adjacency table and read the original vector together for accurate distance calculation. There is also the concept of SECTOR, with the default size is 4096. Each SECTOR only contains 4096 / node_size pieces of vector information. node_size = single vector size + single node adjacency table size.
8.Finally, do a global uniform sampling of 150000 / n, save it, and use it for warmup when searching.
Search
There are 10 search parameters:
index_type: Options include Float/int8/uint8, similar to the first parameter data_type when building an index.
index_prefix_path: Refer to index parameter index_prefix_path.
num_nodes_to_cache: Number of cache hotspots.
num_threads: Number of search threads.
beamwidth: Upper limit of the number of preload points. The system determines if it is set 0.
query_file.bin: Query set file.
truthset.bin: Result set file, "null" means that the result set is not provided, the program calculates it by itself;
K: topk;
result_output_prefix: Path to save search results;
L*: Search parameter list. Multiple values can be added. For each L, statistical information will be given while searching with different L.
Search process:
Load related data: load query set, PQ center point data, codebook data, search starting point and other data, and read index meta.
Use the data set sampled during indexing to do cached_beam_search, count the access times of each point, and load num_nodes_to_cache points with the highest access frequency to the cache.
There is a WARMUP operation by default. Like step 2, this sample data set is also used to do a cached_beam_search.
According to the number of parameters L given, each L will be performed with cached_beam_search again with the query set, and statistics such as recall rate and QPS will be output. The process of warmup and statistics hotspot data is not counted in the query time.
About cached_beam_search:
Find the closest candidate to the query point from the candidate starting point. The PQ distance is used here, and the starting point is added to the search queue.
Start searching:
From the search queue, there are no more than beam_width + 2 unvisited points. If these points are in the cache, add them to the cache hit queue. If they are not hit, add them to the miss queue. Make sure that the size of the miss queue does not exceed beam_width.
Send asynchronous disk access requests to points in the miss queue.
For the points hit by the cache, use the original data and the query data to calculate the exact distance, add to the result queue, and then use PQ to calculate the distance to the neighbor points that have not been visited before adding to the search queue. The length of the search queue is limited by parameters.
Process the cached miss points in step a, similar to step c.
When the search queue is empty, the search ends, and the result queue topk is returned.
Summarize
Although this is a relatively lengthy piece of work, it is overall excellent. The paper and code ideas are clear: divide a number of overlapping buckets through k-means, and then divide the buckets to build a map index, and finally merge the indexes, which is a relatively new idea. As for the memory-based graph index Vamana, it is essentially a randomly initialized version of NSG that can control the trimming granularity. When querying, it makes full use of cache + pipeline, covers up part of the io time, and improves QPS. However, according to the paper, even if the machine condition is not extraordinary, the training time takes up to 5 days, and the usability is relatively low. Optimizations to the training are definitely necessary in the future. From the code perspective, the quality is relatively high and can be directly used in the production environment.
References
[Cong Fu, Chao Xiang, Changxu Wang, and Deng Cai. Fast approximate nearest neighbor search with the navigating spreading-out graphs. PVLDB, 12(5):461 – 474, 2019. doi: 10.14778/3303753.3303754.] (http://www.vldb.org/pvldb/vol12/p461-fu.pdf)
Cong Fu and Deng Cai. GitHub - ZJULearning/efanna: fast library for ANN search and KNN graph construction.
Start Free, Scale Easily
Try the fully-managed vector database built for your GenAI applications.
Try Zilliz Cloud for FreeKeep Reading

Our Journey to 35K+ GitHub Stars: The Real Story of Building Milvus from Scratch
Join us in celebrating Milvus, the vector database that hit 35.5K stars on GitHub. Discover our story and how we’re making AI solutions easier for developers.

Why Deepseek is Waking up AI Giants Like OpenAI And Why You Should Care
Discover how DeepSeek R1's open-source AI model with superior reasoning capabilities and lower costs is disrupting the AI landscape and challenging tech giants like OpenAI.

Enhancing AI Reliability Through Fine-Grained Hallucination Detection and Correction with FAVA
In this blog, we will explore the nature of hallucinations, the taxonomy that provides a framework for categorizing them, the FAVABENCH dataset designed for evaluation, and how FAVA detects and corrects errors.
