Accelerating Similarity Search on Really Big Data with Vector Indexing

From computer vision to new drug discovery, vector similarity search engines power many popular artificial intelligence (AI) applications. A huge component of what makes it possible to efficiently query the million-, billion-, or even trillion-vector datasets that similarity search engines rely on is indexing, a process of organizing data that drastically accelerates big data search. This article covers the role indexing plays in making vector similarity search efficient, different vector inverted file (IVF) index types, and advice on which index to use in different scenarios.
Jump to:
- How does vector indexing accelerate similarity search and machine learning?
- What are different types of IVF indexes and which scenarios are they best suited for?
- FLAT: Good for searching relatively small (million-scale) datasets when 100% recall is required.
- IVF_FLAT: Improves speed at the expense of accuracy (and vice versa).
- IVF_SQ8: Faster and less resource hungry than IVF_FLAT, but also less accurate.
- IVF_SQ8H: New hybrid GPU/CPU approach that is even faster than IVF_SQ8.
- Learn more about Milvus, a massive-scale vector data management platform.
How does vector indexing accelerate similarity search and machine learning?
Similarity search engines work by comparing an input to a database to find objects that are most similar to the input. Indexing is the process of efficiently organizing data, and it plays a major role in making similarity search useful by dramatically accelerating time-consuming queries on large datasets. After a massive vector dataset is indexed, queries can be routed to clusters, or subsets of data, that are most likely to contain vectors similar to an input query. In practice, this means a certain degree of accuracy is sacrificed to speed up queries on really big vector data.
An analogy can be drawn to a dictionary, where words are sorted alphabetically. When looking up a word, it is possible to quickly navigate to a section that only contains words with the same initial — drastically accelerating the search for the input word’s definition.
What are different types of IVF indexes and which scenarios are they best suited for?
There are numerous indexes designed for high-dimensional vector similarity search, and each one comes with tradeoffs in performance, accuracy, and storage requirements. This article covers several common IVF index types, their strengths and weaknesses, as well as performance test results for each index type. Performance testing quantifies query time and recall rates for each index type in Milvus, an open-source vector data management platform. For additional information on the testing environment, see the methodology section at the bottom of this article.
FLAT: Good for searching relatively small (million-scale) datasets when 100% recall is required.
For vector similarity search applications that require perfect accuracy and depend on relatively small (million-scale) datasets, the FLAT index is a good choice. FLAT does not compress vectors, and is the only index that can guarantee exact search results. Results from FLAT can also be used as a point of comparison for results produced by other indexes that have less than 100% recall.
FLAT is accurate because it takes an exhaustive approach to search, which means for each query the target input is compared to every vector in a dataset. This makes FLAT the slowest index on our list, and poorly suited for querying massive vector data. There are no parameters for the FLAT index in Milvus, and using it does not require data training or additional storage.
FLAT performance test results:
FLAT query time performance testing was conducted in Milvus using a dataset comprised of 2 million 128-dimensional vectors.
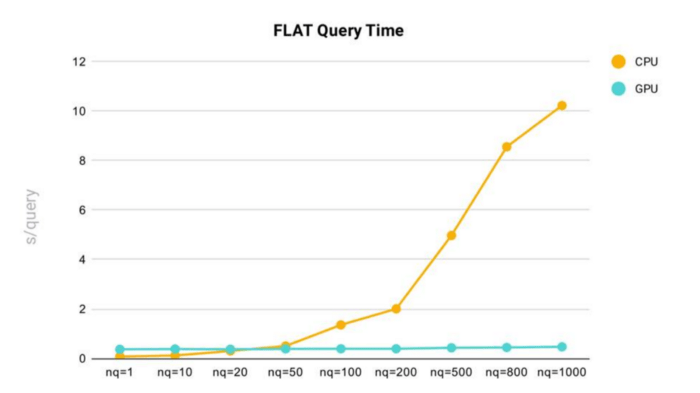 Query time test results for the FLAT index in Milvus.
Query time test results for the FLAT index in Milvus.
Key takeaways:
- As nq (the number of target vectors for a query) increases, query time increases.
- Using the FLAT index in Milvus, we can see that query time rises sharply once nq exceeds 200.
- In general, the FLAT index is faster and more consistent when running Milvus on GPU vs. CPU. However, FLAT queries on CPU are faster when nq is below 20.
IVF_FLAT: Improves speed at the expense of accuracy (and vice versa).
A common way to accelerate the similarity search process at the expense of accuracy is to conduct an approximate nearest neighbor (ANN) search. ANN algorithms decrease storage requirements and computation load by clustering similar vectors together, resulting in faster vector search. IVF_FLAT is the most basic inverted file index type and relies on a form of ANN search.
IVF_FLAT divides vector data into a number of cluster units (nlist), and then compares distances between the target input vector and the center of each cluster. Depending on the number of clusters the system is set to query (nprobe), similarity search results are returned based on comparisons between the target input and the vectors in the most similar cluster(s) only — drastically reducing query time.
By adjusting nprobe, an ideal balance between accuracy and speed can be found for a given scenario. Results from our IVF_FLAT performance test demonstrate that query time increases sharply as both the number of target input vectors (nq), and the number of clusters to search (nprobe), increase. IVF_FLAT does not compress vector data however, index files include metadata that marginally increases storage requirements compared to the raw non-indexed vector dataset.
IVF_FLAT performance test results:
IVF_FLAT query time performance testing was conducted in Milvus using the public 1B SIFT dataset, which contains 1 billion 128-dimensional vectors.
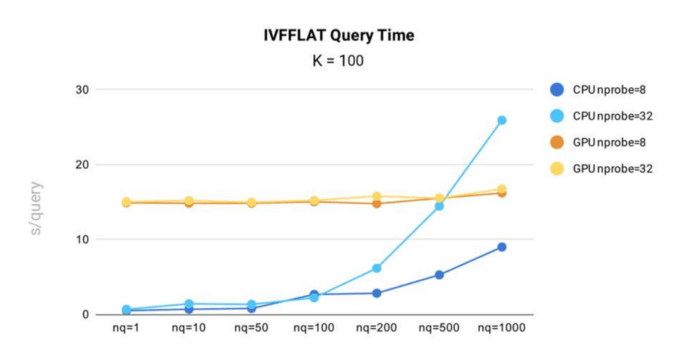 Query time test results for IVF_FLAT index in Milvus.
Query time test results for IVF_FLAT index in Milvus.
Key takeaways:
- When running on CPU, query time for the IVF_FLAT index in Milvus increases with both nprobe and nq. This means the more input vectors a query contains, or the more clusters a query searches, the longer query time will be.
- On GPU, the index shows less time variance against changes in nq and nprobe. This is because the index data is large, and copying data from CPU memory to GPU memory accounts for the majority of total query time.
- In all scenarios, except when nq = 1,000 and nprobe = 32, the IVF_FLAT index is more efficient when running on CPU.
IVF_FLAT recall performance testing was conducted in Milvus using both the public 1M SIFT dataset, which contains 1 million 128-dimensional vectors, and the glove-200-angular dataset, which contains 1+ million 200-dimensional vectors, for index building (nlist = 16,384).
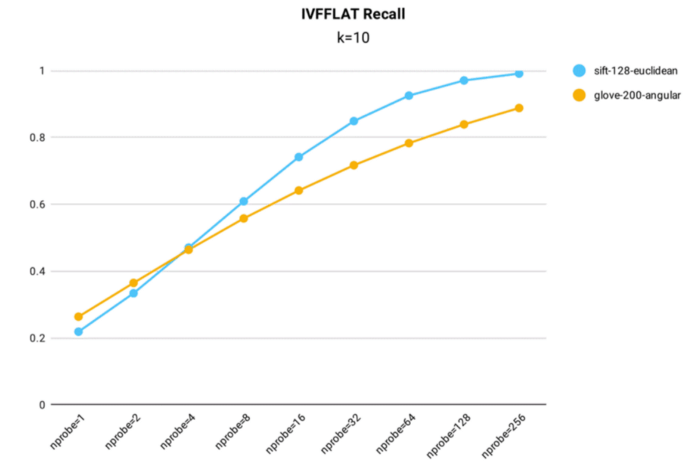 Recall rate test results for the IVF_FLAT index in Milvus.
Recall rate test results for the IVF_FLAT index in Milvus.
Key takeaways:
- The IVF_FLAT index can be optimized for accuracy, achieving a recall rate above 0.99 on the 1M SIFT dataset when nprobe = 256.
IVF_SQ8: Faster and less resource hungry than IVF_FLAT, but also less accurate.
IVF_FLAT does not perform any compression, so the index files it produces are roughly the same size as the original, raw non-indexed vector data. For example, if the original 1B SIFT dataset is 476 GB, its IVF_FLAT index files will be slightly larger (~470 GB). Loading all the index files into memory will consume 470 GB of storage.
When disk, CPU, or GPU memory resources are limited, IVF_SQ8 is a better option than IVF_FLAT. This index type can convert each FLOAT (4 bytes) to UINT8 (1 byte) by performing scalar quantization. This reduces disk, CPU, and GPU memory consumption by 70–75%. For the 1B SIFT dataset, the IVF_SQ8 index files require just 140 GB of storage.
IVF_SQ8 performance test results:
IVF_SQ8 query time testing was conducted in Milvus using the public 1B SIFT dataset, which contains 1 billion 128-dimensional vectors, for index building.
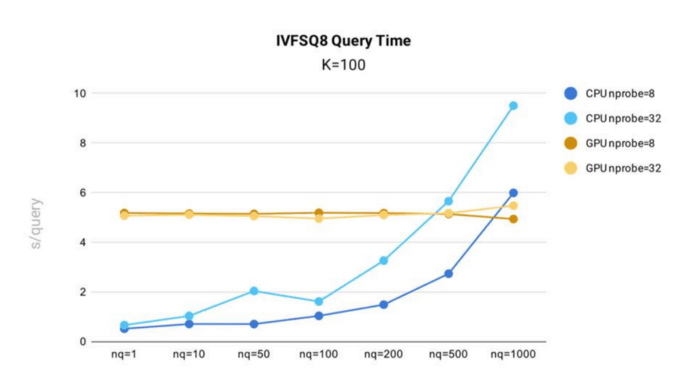 Query time test results for IVF_SQ8 index in Milvus.
Query time test results for IVF_SQ8 index in Milvus.
Key takeaways:
- By reducing index file size, IVF_SQ8 offers marked performance improvements over IVF_FLAT. IVF_SQ8 follows a similar performance curve to IVF_FLAT, with query time increasing with nq and nprobe.
- Similar to IVF_FLAT, IVF_SQ8 sees faster performance when running on CPU and when nq and nprobe are smaller.
IVF_SQ8 recall performance testing was conducted in Milvus using both the public 1M SIFT dataset, which contains 1 million 128-dimensional vectors, and the glove-200-angular dataset, which contains 1+ million 200-dimensional vectors, for index building (nlist = 16,384).
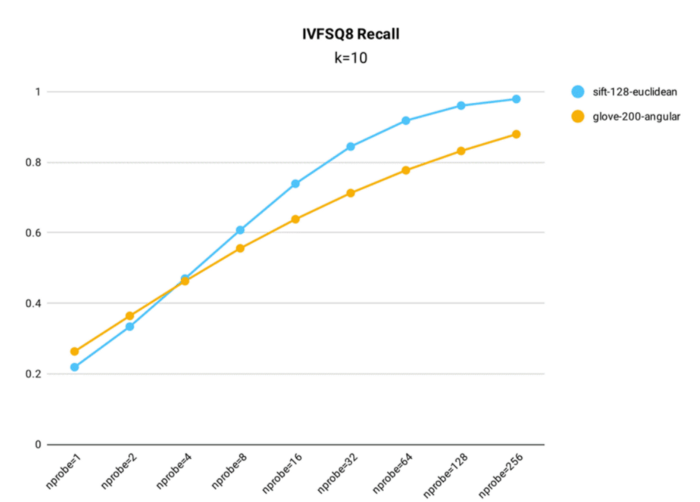 Recall rate test results for IVF_SQ8 index in Milvus.
Recall rate test results for IVF_SQ8 index in Milvus.
Key takeaways:
- Despite compressing the original data, IVF_SQ8 does not see a significant decrease in query accuracy. Across various nprobe settings, IVF_SQ8 has at most a 1% lower recall rate than IVF_FLAT.
IVF_SQ8H: New hybrid GPU/CPU approach that is even faster than IVF_SQ8.
IVF_SQ8H is a new index type that improves query performance compared to IVF_SQ8. When an IVF_SQ8 index running on CPU is queried, most of the total query time is spent finding nprobe clusters that are nearest to the target input vector. To reduce query time, IVF_SQ8 copies the data for coarse quantizer operations, which is smaller than the index files, to GPU memory — greatly accelerating coarse quantizer operations. Then gpu_search_threshold determines which device runs the query. When nq >= gpu_search_threshold, GPU runs the query; otherwise, CPU runs the query.
IVF_SQ8H is a hybrid index type that requires the CPU and GPU to work together. It can only be used with GPU-enabled Milvus.
IVF_SQ8H performance test results:
IVF_SQ8H query time performance testing was conducted in Milvus using the public 1B SIFT dataset, which contains 1 billion 128-dimensional vectors, for index building.
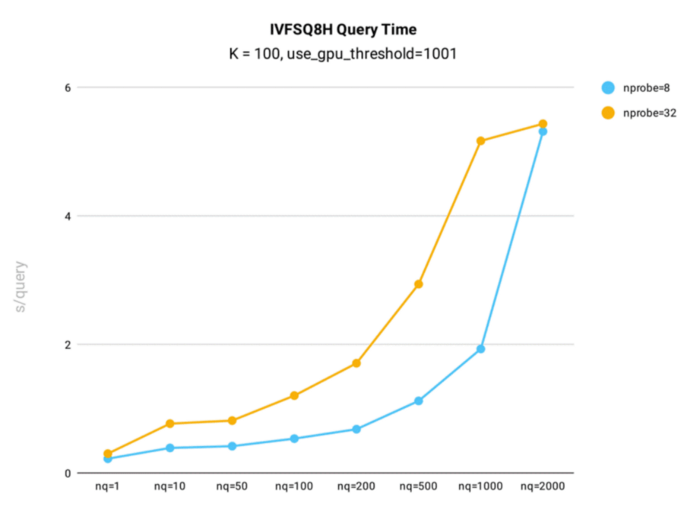 Query time test results for IVF_SQ8H index in Milvus.
Query time test results for IVF_SQ8H index in Milvus.
Key takeaways:
- When nq is less than or equal to 1,000, IVF_SQ8H sees query times nearly twice as fast as IVFSQ8.
- When nq = 2000, query times for IVFSQ8H and IVF_SQ8 are the same. However, if the gpu_search_threshold parameter is lower than 2000, IVF_SQ8H will outperform IVF_SQ8.
- IVF_SQ8H’s query recall rate is identical to IVF_SQ8’s, meaning less query time is achieved with no loss in search accuracy.
Learn more about Milvus, a massive-scale vector data management platform.
Milvus is a vector data management platform that can power similarity search applications in fields spanning artificial intelligence, deep learning, traditional vector calculations, and more. For additional information about Milvus, check out the following resources:
- Milvus is available under an open-source license on GitHub.
- Additional index types, including graph- and tree-based indexes, are supported in Milvus. For a comprehensive list of supported index types, see documentation for vector indexes in Milvus.
- To learn more about the company that launched Milvus, visit Zilliz.com.
- Chat with the Milvus community or get help with a problem on Slack.
Methodology
Performance testing environment
The server configuration used across performance tests referenced in this article is as follows:
- Intel (R) Xeon (R) Platinum 8163 @ 2.50GHz, 24 cores
- GeForce GTX 2080Ti x 4
- 768 GB memory
Relevant technical concepts
Although not required for understanding this article, here are a few technical concepts that are helpful for interpreting the results from our index performance tests:
 Blog_Accelerating Similarity Search on Really Big Data with Vector Indexing_8.png
Blog_Accelerating Similarity Search on Really Big Data with Vector Indexing_8.png
Resources
The following sources were used for this article:
- “Encyclopedia of database systems,” Ling Liu and M. Tamer Özsu.
What's Next
Continue to read Accelerating Similarity Search on Really Big Data with Vector Indexing: Part II.
Start Free, Scale Easily
Try the fully-managed vector database built for your GenAI applications.
Try Zilliz Cloud for FreeKeep Reading

Our Journey to 35K+ GitHub Stars: The Real Story of Building Milvus from Scratch
Join us in celebrating Milvus, the vector database that hit 35.5K stars on GitHub. Discover our story and how we’re making AI solutions easier for developers.

Why Deepseek is Waking up AI Giants Like OpenAI And Why You Should Care
Discover how DeepSeek R1's open-source AI model with superior reasoning capabilities and lower costs is disrupting the AI landscape and challenging tech giants like OpenAI.

RocketQA: Optimized Dense Passage Retrieval for Open-Domain Question Answering
RocketQA is a highly optimized dense passage retrieval framework designed to enhance open-domain question-answering (QA) systems.
